Democratizing Biology: How Affordable AI and Automation are Empowering Labs
By Mike Fussell, Life Sciences Product Manager
Last updated on Nov. 28, 2024
Accelerating Research and Reducing Costs for Academic and Corporate Labs
Introduction
AI is transforming the field of biology from fundamental research to applied disciplines like drug discovery. It is empowering research labs to improve throughput and consistency, and startups to scale faster and more affordably to challenge industry incumbents with new and exciting technologies.
AI-based image processing of widefield fluorescence images can yield images with fine details that go beyond the diffraction limit, potentially eliminating the need for conventional super-resolution imaging systems which are complex and expensive. At the same time, AI systems can recognize complex patterns in highly variable data and reliably distinguish between subtly different classes of cells or based on features which could easily go unnoticed by human viewers.
Automation is a key capability for research groups and companies looking to get the most out of AI. Automated microscopes make it possible to capture more images, more consistently in less time. This enables both the rapid acquisition of large high-quality AI training datasets, and the deployment of AI models trained on those datasets for applications like drug discovery.
This article gives a brief overview of how the combination of AI and automation can be used to empower academic and corporate labs to do more with less.
Democratizing Super-Resolution Imaging
Until recently, the diffraction limit placed a fundamental limit on the amount of detail that could be resolved by light microscopy. By exploiting the unique properties of certain fluorophores, deterministic and stochastic methods capable of generating images with resolutions greater than the diffraction limit were developed. While effective, techniques like STED, STORM, TIRF and lattice light sheet all require complex and expensive instrumentation which places them beyond the reach of many academic and corporate research labs.
The application of convolutional neural networks to widefield fluorescence microscopy to generate super-resolution images has the potential to democratize super-resolution microscopy. For many labs, this can reduce or eliminate the need for expensive, and complex imaging systems while still delivering high quality, trustworthy data.
Advantages and Limitations of AI Enhanced Images
AI image enhancement builds on the strengths of earlier computational deconvolution approaches employed by software tools such as Huygens. AI enhancement of widefield fluorescence images yields greater resolution and reduced noise. It also helps to overcome one of the major weaknesses of traditional widefield imaging, even going so far as to turn it into a strength! Many super-resolution imaging techniques achieve excellent SNR by means of extremely thin optical sectioning. By minimizing the amount of excitation light that reaches out of focus fluorophores, these techniques prevent the background from being washed out by emitted light from out of focus regions. AI enhancement can remove the unwanted light from out of focus regions to deliver images with much greater SNR (Fig. 1).
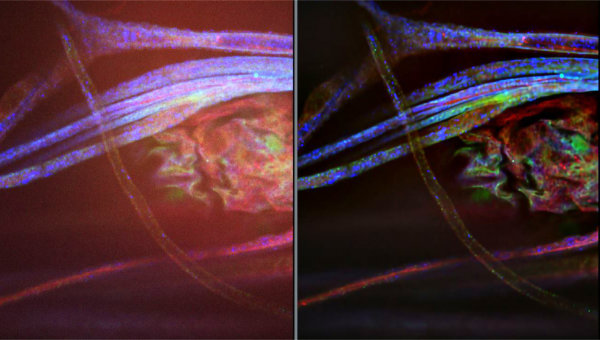
The thin optical sectioning employed by super-resolution imaging methods can also create challenges for samples which are thicker, or have regions of interest more than 200 nm from the cover slip or wave guide in the case of TIRF. This shallow depth of field (DOF) can be problematic for imaging cells in microfluidic devices where only cells on one surface of a channel may be observed. The greater DOF of widefield images can eliminate the need for Z-stacking when paired with AI enhancement to sharpen mildly out of focus areas, while eliminating unwanted signal from fluorophores which are further out of the focal plane.
A concern often raised about AI enhanced images is the potential introduction of unwanted artifacts which would reduce the confidence level of decisions made based on them. Anyone who has used ChatGPT is aware of how some AI systems can invent details which seem reasonable at first, but turn out to be incorrect when checked. Fortunately, methods based on supervised training with experimentally acquired super-resolution images paired with Generative Adversarial Networks (GANs) have proven highly reliable when compared against the ground truth3.
Get Started with AI Image Enhancement Today
VisiView4 is a commercial software package from Visitron systems which combines automated image acquisition with AI enhancement and AI image analysis. This software is fully compatible with the Nucleus platform and is optimized to deliver highly reliable results at the fastest possible throughput.
For groups on a tight budget, DeconvolutionLab 21 is an open source package which works in conjunction with FIJI and μManager. This package does not require any prior knowledge about the sample or the optical system or the tuning of any parameters for optimization. While this package is free to download and does not require any license fees, its reliance on μManager, infrequent updates and the absence of any formal support should be considered.
Train and Deploy AI systems at scale with Automation
In addition to super-resolution enhancement, AI systems excel at recognizing complex patterns in highly variable data. This makes AI ideal for the analysis of high content imaging where many different phenotypes are possible. This ability is mandatory for managing the complexity inherent in multi-omics systems.
Large, high-quality datasets are the foundation of any AI-based screening or discovery program. The high value of a unique dataset has been clearly demonstrated by AstraZeneca’s recent round of AI focused startup acquisitions. These were not driven by the acquired companies having unique “killer algorithms” for in silico drug discovery, but rather for AstraZenica to gain access to their unique datasets.
The first step in building a reliable AI-based cell image analysis system is training. This requires the acquisition of a high-quality training dataset. Modular and low cost equipment such as from the Zaber Nucleus Microscopy platform are an invaluable tool for building those datasets quickly. The Nucleus microscopy platform deliver in three key areas:
Maximizing Throughput
Automation delivers greatly improved throughput compared to manual processes. An automated microscope can capture more images in less time. With automated systems it is possible to scan entire 96 well plates in a few seconds. To achieve this level of throughput requires both hardware and software to be optimized for high-speed automation. Zaber provides a comprehensively documented API in eight popular languages including Python, C# and C++ which streamlines development of custom software or integration into existing AI and machine learning workflows. These APIs enable direct control of all functions of the microscope, yielding as much as a 2x increase in throughput compared to less highly optimized software like μManager.
While it is beyond the scope of this article, the Zaber content library has more detailed guides to maximizing your imaging throughput by optimizing your protocol, applying input shaping to minimize the combined move and settle times, and synchronizing a TDI line-scan camera to a high-speed linear motor stage.
Zaber's HL04 laser autofocus module can greatly improve imaging throughput by eliminating the need for image-based autofocus techniques. In addition to the much greater speed of focusing, the long IR wavelength of the autofocus sensor does not bleach fluorophores ensuring they are at the brightest, so exposures can be shorter.
Achieving Consistency
The more complex the problem and the more variability there is in the source data, the larger the training dataset will need to be to provide sufficient coverage of the range of cases the resulting AI system is expected to encounter. Zaber’s Nucleus automated microscopy platform, is an ideal tool for automating the acquisition of training data. Highly repeatable XY and focus stage movement ensures the sample is positioned consistently. Reliable and fast automation of focus is made possible with the HL04 laser autofocus sensor. High-speed stages help to minimize the influence of time-dependent effects, such temperature fluctuations and evaporation to deliver consistent results over an entire microplate. Highly stable epi-illuminator, like the Zaber MLR3, ensures illumination intensity remains consistent between the first well and the last.
Maximizing Versatility
The field of AI is advancing rapidly, as are many supporting technologies like microfluidics and 3D cell culture. Nucleus’ modular architecture makes it easy to build systems which have the capabilities required to automate novel techniques. This opens the door to applying AI where it might not have been possible previously. Nucleus systems can also be reconfigured to evolve as your research projects or processes change over time.
Software is a critical yet often overlooked dimension of versatility. The Zaber Motion Library API makes it easy to integrate all the features of a Nucleus microscope system into your AI workflow. This enables autonomously acquisition of complex multidimensional datasets to train your AI model or to deploy that trained model to tasks like screening large libraries.
Summary
AI and automated widefield fluorescence microscopy are a powerful combination. Together, these technologies form a feedback loop with AI image enhancement revealing a greater amount of meaningful information from widefield images, eliminating the need for conventional super-resolution imaging systems, while automated image acquisition makes it possible to train and deploy AI-based models for image analysis. These tools enable both academic and corporate research groups to achieve amazing results with a fraction of the resources that would otherwise be required.
References
- https://github.com/Biomedical-Imaging-Group/DeconvolutionLab2
- Sage, Daniel, et al. "DeconvolutionLab2: An open-source software for deconvolution microscopy." Methods 115 (2017): 28-41
- Wang, Hongda, et al. "Deep learning enables cross-modality super-resolution in fluorescence microscopy." Nature methods 16.1 (2019): 103-110.
- https://www.visitron.de/products/visiviewr-software.html